ARTEMIS is the name of a new computational method with the potential to significantly advance our understanding of RNA, a key molecule involved in various cellular functions. The algorithm was developed by scientists from the International Institute of Molecular and Cell Biology in Warsaw.
RNA is not just a simple chain of nucleotides that carries genetic information. RNA molecules also have other biological functions, often requiring the formation of complex three-dimensional structures, much like proteins do. However, comparing these intricate shapes of RNA structures has proven challenging, especially when the molecules differ in nucleotide sequences.
Recent advances in structural biology now provide scientists with tools to study RNA 3D structures more effectively and to better understand how these structures enable RNA to function within living organisms. If two RNA molecules have similar structures, a hypothesis can be proposed and tested that they may also share similar functions.
'In 2022, the team designed a new algorithm called ARTEM, based on the idea that in the best possible spatial overlay of two RNA structures, at least one nucleotide from each structure would be nearly ideally superimposed on its counterpart', a press release from the International Institute of Molecular and Cell Biology (IIMCB) in Warsaw says.
The next step was the development of ARTEMIS, to enable comparisons of entire RNA molecular structures and determine which nucleotides in one sequence correspond to nucleotides in another.
‘[Eugene F.] Baulin and collaborating team members conducted preliminary tests and found that ARTEMIS works very well - and for many RNA molecules, even better than the best tools previously developed by other researchers', the press release continues.
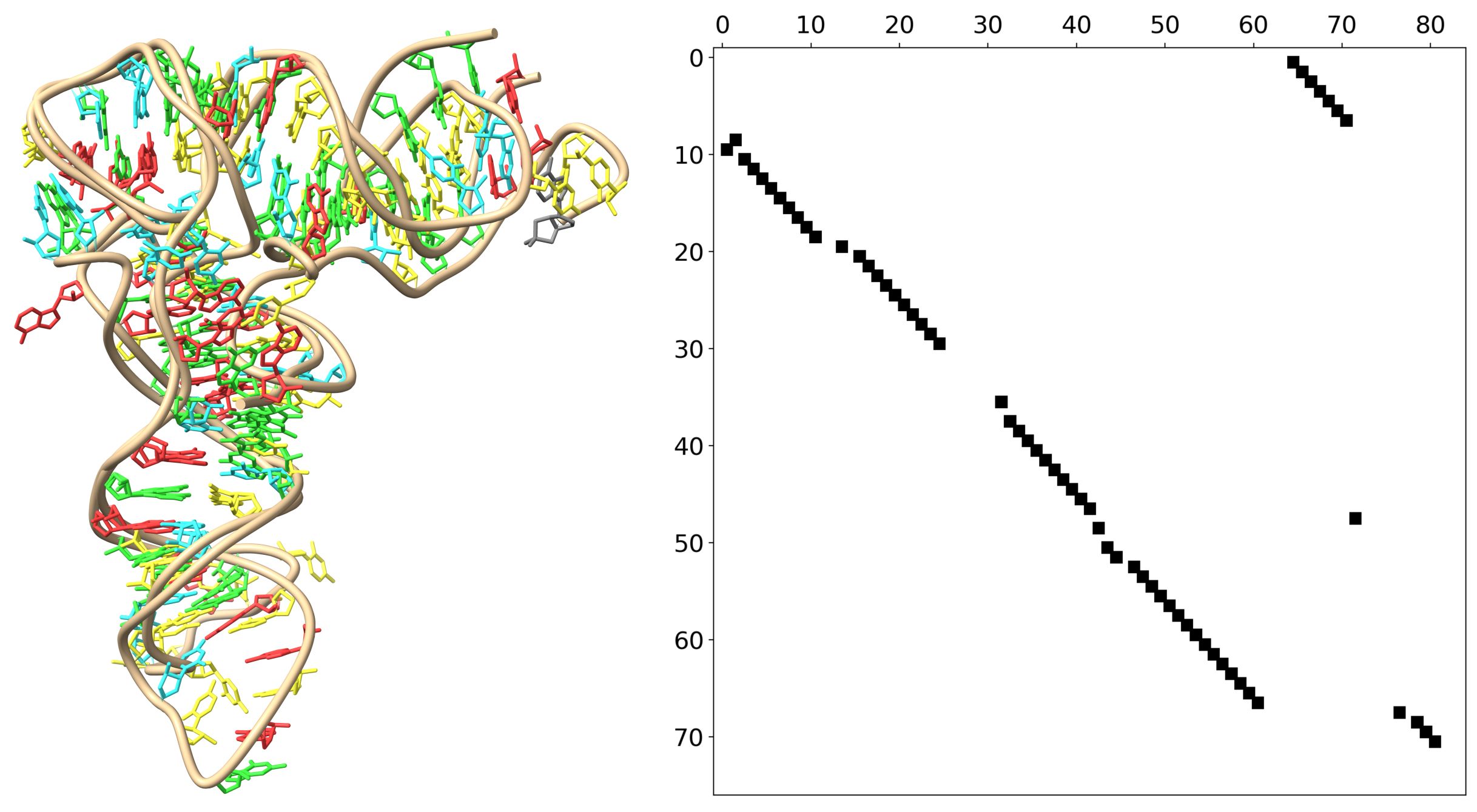
By comparing RNA structures, ARTEMIS can accurately determine which nucleotides correspond to each other. This confirms that the research assumption of identifying structural motifs by aligning individual nucleotides and checking how the remaining nucleotides relate to each other yields excellent results in comparing the spatial structures of entire RNA molecules.
ARTEMIS was designed to analyse RNA's three-dimensional shapes, even in cases where the sequences differ significantly, including changes in nucleotide order. Current tools often struggle with such a task. 'In contrast, ARTEMIS operates without restrictions on the order of nucleotides in the sequences being compared, allowing scientists to compare RNA structures more precisely and effectively', the researchers say.
They add that the new method may interest researchers in structural bioinformatics, who analyse structural databases, classify known structures, and predict the structures of RNA molecules that have not yet been determined. It is also a valuable tool for structural biologists who experimentally determine new structures of nucleic acids, both RNA and DNA. These scientists can use ARTEMIS to search structural databases for similar structures or motifs that may provide insights into biological functions.
Artemis can also contribute to the improved design of new functional RNA molecules and molecules that interact with RNA, applicable in biotechnology and medicine. According to Baulin, practically every introduction of a truly innovative invention into everyday use stems from earlier basic research - such as the one that led to the development of the ARTEMIS method.
'What excites me most is how one simple assumption about RNA structure led to the development of powerful tools that allow us to make new discoveries', says Eugene F. Baulin, According to the researcher, it shows how 'a deep understanding of data can lead to a simple solution that outperforms more complex approaches'.
'With the success of the AlphaFold artificial intelligence algorithm in predicting protein structures, RNA structure prediction has now become the next holy grail of computational biology, and each new tool, like ARTEMIS, brings us a little closer to that solution', says Baulin from the Laboratory of Bioinformatics and Protein Engineering at the International Institute of Molecular and Cell Biology in Warsaw. (PAP)
Urszula Kaczorowska
uka/ bar/ kap/
tr. RL













