Inside human cells, the helical DNA strand twists further and is completely different from the familiar double helix. At very small scales, the behaviour of DNA is determined by elastic forces. Scientists show how to study the elastic properties of DNA using hydrodynamic measurements. Discovering the shapes of DNA mini-circles was described by an international research team with the participation of Polish scientists.
Researchers from the Faculty of Physics of the University of Warsaw, the Polish Academy of Sciences, Baylor College of Medicine, Rice University, University of Montana and University of Lethbridge studied DNA geometry. They proposed new ways to study active DNA, including a way to study the mechanics of over-twisting and overcoming the problem of its relaxation. The solutions are DNA mini-circles. Polish researchers developed a theoretical approach. They assumed that DNA could be modelled as a homogeneous elastic bundle behaving like a miniature rubber band - which was confirmed by experiments. The research results were published in the journal Nucleic Acids Research.
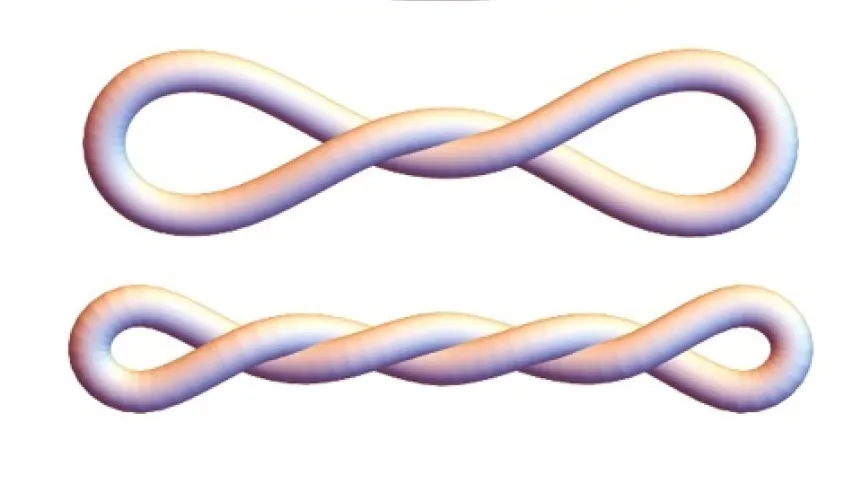
Inside the cells, where the DNA performs its most important functions, it is subjected to replication and transcription machinery. This results in transient under- and over-twisting of that helical double-strand. Such shapes can also be observed at the human scale, for example by grabbing an end of a string (or headphones) and twisting it. After a few rotations it coils up, a term used for DNA is 'supercoiling'.

Studying the under-twisting and over-twisting mechanics is particularly challenging. Most research on the topic of DNA geometry is conducted with short linear fragments. Even if they were twisted the ends, they would spontaneously relax the twist. 'For illustration, twist a shoelace - if you let go of either end, the shoelace just becomes straight (untwisted) again,’ says Radost Waszkiewicz, the leading co-author of the paper and a doctoral candidate at the Faculty of Physics of the University of Warsaw.
The twist relaxation does not occur if the ends of the DNA segment are attached together - this way the twist is 'locked in' within the loop and cannot 'escape' through the ends. A group of researchers from the US, led by Professor Lynn Zecheidrich, figured out how to do just that using small (336 base pair long) DNA minicircles.
DNA LIKE A MINIATURE RUBBER BAND - POLISH CONTRIBUTION TO RESEARCH
The theoretical approach was developed by Radost Waszkiewicz, Professor Maciej Lisicki, and Professor Piotr Szymczak at the Faculty of Physics, University of Warsaw, in collaboration with Professor Maria Ekiel-Jeżewska at the Institute of Fundamental Technology Research of the Polish Academy of Sciences.
'We assumed that the DNA can be modelled as a uniform elastic beam, behaving like a miniature rubber band. For a given amount of twist, we determined the shapes that minimise the total elastic energy, and these are the shapes that we see in experiments! We then used these shapes to calculate their hydrodynamic properties, that is to see how fast they diffuse and sediment in solution,’ says Waszkiewicz.
To understand the dynamics of DNA, or generally small objects in aqueous solvents, we need to explore the mechanics of very viscous fluids. 'Microscale flows are dominated by viscous effects and inertia is completely negligible. Our modelling approach relies on the mathematical properties of these so-called Stokes flows,’ says Lisicki, who was a co-investigator specialising in modelling hydrodynamic interactions.
SUPER TWISTED MINI-CIRCLES EXCITE RESEARCHERS
When the DNA is under or over-twisted more and more before closing it in a mini-circle, its shape becomes more and more compact. These mini-circles with precise defined degrees of twist were investigated in various ways. Their shapes were explored by the Zechiedrich group (Dr. Jonathan Fogg and Dr. Daniel Catanese) using biochemical and biophysical methods, including an electron microscopy technique capable of capturing 3D images of DNA nanoparticles.
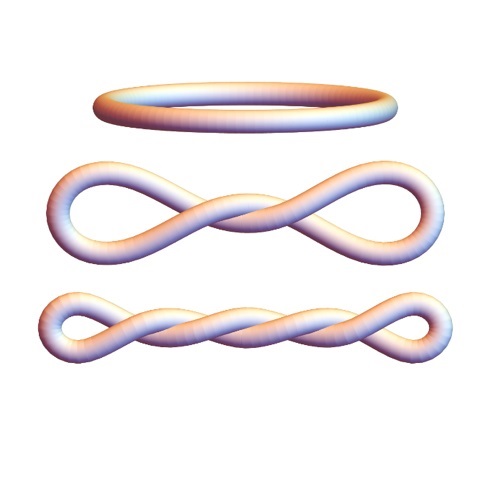
The group led by Professor Borries Demeler at the University of Montana and University of Lethbridge used analytical ultracentrifugation (AUC), a very precise 'first principles' (meaning the read out is direct; no need to compare to reference to infer) method to measure how quickly the molecules sediment, which depends on their diffusion and sedimentation coefficients. Maduni Ranasinghe, a doctoral candidate at Lethbridge, performed AUC measurements for different supercoiling density of the mini-circles. Increasing the compactness of the DNA molecules was shown to increase the diffusion coefficient.
'We continue to be amazed by how different supercoiled DNA is from the form we know a lot about - the so-called +B-form+ DNA double helix that you see depicted in sculpture, architecture, and art. Active DNA looks very different from that form and we are so excited to continue to discover the ways access to the primary genetic code is regulated,’ says Professor Lynn Zechiedrich.
PAP - Science in Poland
kol/ zan/ kap/
tr. RL













