
Well-preserved DNA of several hundred species of bacteriophages from over a thousand years ago - viruses that infect bacteria - have been found in samples of human faeces and in the guts of an ancient mummy. It turns out that the genome of one of the species is almost the same as that of modern viruses. Why has the evolution of this species halted for so long?
The results of research conducted by an international team led by Polish scientists, published in Nature Communications, change the perception of the 'arms race' between viruses and their bacterial hosts.
'What we have found seems to contradict our previous knowledge about bacteriophages,’ the first author of the study Piotr Rozwalak from the Adam Mickiewicz University in Poznań tells PAP. The research was conducted as part of his master's thesis.
In order to reproduce, viruses must enter the host cell and make it copy their genetic material. There are viruses that attack plant, human and animal cells. Viral infections also threaten bacterial cells. Such bacteria-specific viruses are bacteriophages (or for short: phages).
Both bacteria and bacteriophages inhabit the human digestive system. Some are beneficial or neutral to human health, others not necessarily so. Their DNA can be found in faeces samples or material collected from the intestines.
Bioinformatics expert and co-author of the paper, Dr. Andrzej Zieleziński from the Adam Mickiewicz University, says that thanks to the achievements of metagenomics, it is possible to sequence and read DNA fragments found in a sample of biological material taken from any environment (e.g. soil or the human digestive system). These cut 'puzzle pieces' can be assembled into larger fragments, including entire genomes, and compared with the already known genomes of various organisms.
This makes it possible to sequence millions of fragments of DNA strands contained in a sample of material, even if the sample is thousands or millions of years old but perfectly preserved.
RECYCLING OF PALAEOFAECES
In this way, teams from all over the world examined human faeces from over 1,000 years ago, preserved in desert caves in Mexico and the United States (Arizona and Utah). Samples were also taken from the intestines of a 5,000-year-old human mummy found in the Alps, or in a salt mine dating back to the Bronze Age.
Interestingly, data from these studies were publicly available. To gain access to data on fossil DNA contained in palaeofaeces, there was no need to conduct expensive searches, archaeological research and genome sequencing; it was enough to analyse publicly available databases and look for different information than what their authors were initially looking for.
'We have carried out such data recycling,’ says Piotr Rozwalak.
While previous research by other teams focused on centuries-old bacteria of the intestinal flora, the team led by Polish scientists decided to focus on the genome of ancient bacteriophages inhabiting the human digestive system. 'We thus took a step into a new field of knowledge, viromics, i.e. the study of viruses present in various environmental samples. We were among the first to examine the samples for bacteriophages. We opened the door to a completely new sub-field, which is fossil viromics, the study of viruses from the past,’ says Rozwalak.
ARMS RACE
The scientists decided to check how quickly the genome of bacterial viruses had changed over the last few centuries. They assumed that at least some fossil bacteriophages would be similar to modern ones. They realized that the obstacle could be the evolutionary process that would erase any traces of similarity between subsequent generations of viruses. It's about a constant arms race between bacteria and viruses, which can quickly change both sides beyond recognition.
Bacteriophages often contribute to the death or weakening of bacteria. It is therefore usually in the interest of bacteria to produce effective defence mechanisms against their pathogens. If one bacterium finds a way to defend itself against infection, it can pass on this genetic 'software update' to its neighbours. And then only those viruses that find a new breach in the host's defences will multiply. So it seemed that bacteriophages and their hosts were always evolving very rapidly.
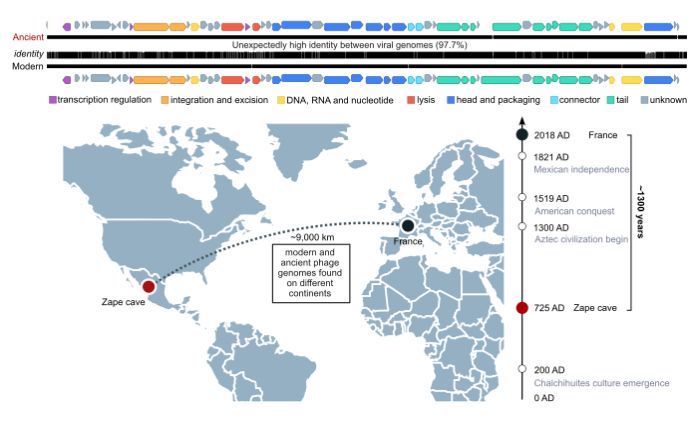
And here came a surprise. Researchers found the genomes of almost 300 species of bacteriophages, which were partly similar to those found today. One of the viruses, however, was unique: its genomic sequence was 97.7 percent similar to the genome of a modern bacteriophage. That virus is called Mushuvirus mushu. It infects anaerobic intestinal bacteria that have a positive impact on our health, including the Faecalibacterium genus.
INVARIABLY
What made the descendants of the virus look the same as their ancestors from 1,300 years ago? This indicates 'a long-term relationship between the prophage and its host,’ the authors of the publication write.
Rozwalak says: 'Our virus is incorporated into the bacterial genome. And for some reason the bacteria does not defend itself against it. Our hypothesis is that there is a mutualistic relationship: each party somehow benefits from the other party's presence - it receives something and wants to cooperate. So, based on these studies, we can guess that sometimes bacteria can cooperate with phages and benefit from their presence.’
Dr. Zieleziński additionally suggests that the stability of the virus may also be due to its ability to infect as many as eight different species of bacteria. And this proves that viruses are very versatile. However, in such a situation, significant changes in the viral DNA may lead the loss of ability to infect one of the possible hosts. So the virus has no interest in evolving.
SEARCHING THE FAECES
Dr. Zieleziński explains that when researching fossil DNA, one should be extremely sceptical because there is a possibility of accidental contamination of samples with modern DNA. Therefore, the authors conducted a series of rigorous tests to verify the authenticity of the fossil virus. The test results clearly showed that Mushuvirus mushuwas as old as the tested sample.
'We have conducted the world's first reconstruction of the genomes of fossil bacteriophages in samples representing the human intestinal environment from up to 5,000 years ago and compiled a catalogue of these viruses, shedding some light on previously unknown aspects of the history of the intestinal microworld. While doing that, we discovered one species of virus that not only had survived for centuries in the human intestine, but also had retained its unchanged form for at least 1,300 years,’ Dr. Zieleziński says.
PAP - Science in Poland, Ludwika Tomala
lt/ agt/ kap/
tr. RL













