Scientists from the University of Warsaw, in collaboration with colleagues from Japan, are conducting research on riboswitches - RNA segments that control the production of proteins in bacteria. In the future, their research may result in new strategies for combating antibiotic resistance in microorganisms.
Scientists from the University of Warsaw, in collaboration with colleagues from Japan, are conducting research on riboswitches - RNA segments that control the production of proteins in bacteria. In the future, their research may result in new strategies for combating antibiotic resistance in microorganisms.
According to the website of the University of Warsaw, Professor Joanna Trylska and Piotr Chyży from the Centre of New Technologies of the University of Warsaw and Dr. Marta Kulik from the Faculty of Chemistry of the same university, in collaboration with Professor Yuji Sugita's group from the Japanese research institute Rikagaku Kenkyūjo (RIKEN), performed molecular dynamics simulations for riboswitch-antibiotic systems. They described the results of their research in the journal Proceedings of the National Academy of Sciences (https://doi.org/10.1073/pnas.2317197121).
They performed all simulations using supercomputers, including those located at the University of Warsaw.
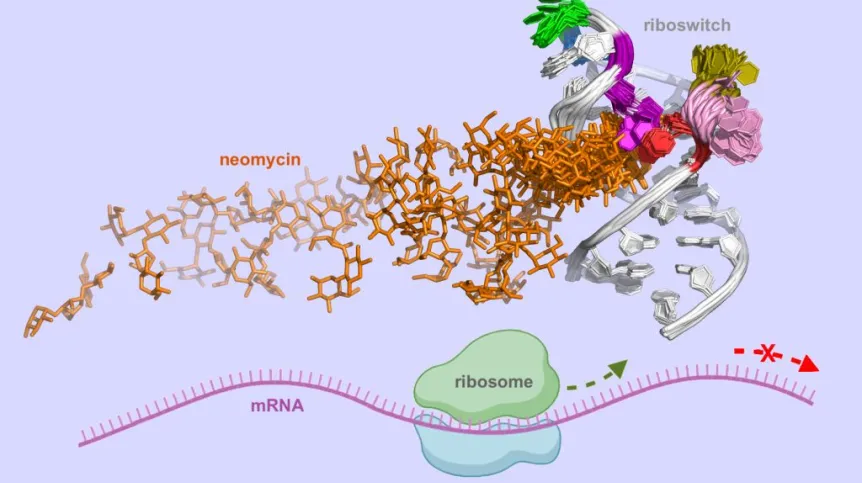
Riboswitches are segments of messenger RNAs (mRNAs) located in front of the protein-coding part of the mRNA. They switch off protein production when they bind to specific small molecules, usually metabolites. Influencing the state of riboswitches could provide control over the production of specific proteins essential for cell life.
'Naturally occurring riboswitches could be targets for new antibiotics. Designing molecules that block these switches in bacteria could lead to the death of bacterial cells,' says Professor Trylska from the Centre of New Technologies at the University of Warsaw.
However, riboswitches are not present in all cells. That is why the researchers focused on their synthetic counterparts, which could be incorporated into mRNA to gain control over the production of particular proteins.
Designing riboswitches is challenging because it requires predicting how their structure will change upon binding to a specific molecule. 'Even if a molecule binds tightly to a riboswitch, it does not always ensure that the riboswitch will function as expected in living cells,’ says Trylska.
Scientists from the University of Warsaw and RIKEN investigated a small synthetic riboswitch that binds neomycin. They developed a method and tools to describe the association pathway of neomycin to the riboswitch. The results of their simulations can help design synthetic riboswitches and molecules that control their dynamics and structure.
'Our work shows how the crucial steps of the interaction of neomycin with the riboswitch can be optimised through mutations to enhance riboswitch regulatory effectiveness in cells. The results highlight key dynamic and structural differences along the binding pathway for the different experimentally reported mutants, unravelling the effects of mutations on the riboswitch activity,’ says Trylska.
The researchers hope that their methodology can facilitate the drug design process by helping to select the most promising molecule-riboswitch pairs for further experimental validation. They would like their results to contribute to accelerating the design of controllable RNA fragments as a potential strategy in the fight against antibiotic resistance. (PAP)
Katarzyna Czechowicz
kap/ bar/
tr. RL













