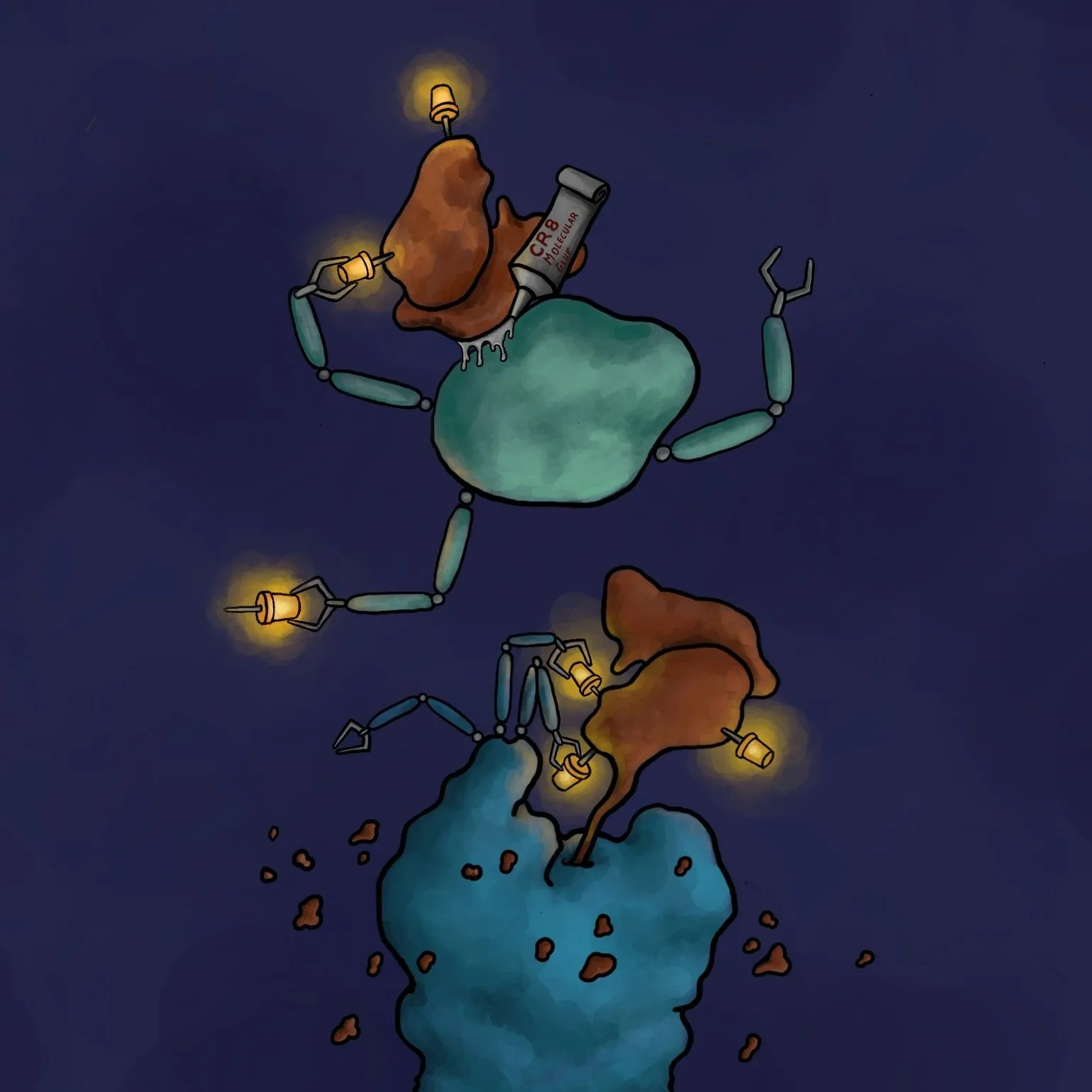
A compound that acts as a glue combining a malfunctioning protein with another protein, which neutralizes it, carrying it to our cellular 'molecular rubbish bin' has been discovered by scientists including Polish researchers.
This is a step on the road to identifying further molecular adhesives that can be used, for example, in targeted cancer therapy.
When a protein in a body doesn't work as it should, it can cause a disease, for example cancer. Drugs used in the treatment of various diseases usually affect selected pathogenic proteins. Molecular adhesives are an innovative mechanism of action of drugs that makes it possible to neutralize even those proteins that were previously beyond the reach of pharmacology.
In an interview with PAP, two Polish scientists, Zuzanna Kozicka from the Friedrich Miescher Institute for Biomedical Research in Basel and Mikołaj Słabicki from the Broad Institute of MIT and Harvard in Cambridge, Dana-Farber Cancer Institute in Boston and the German Cancer Research Center in Heidelberg talked about their discovery of a new type of 'molecular glue’.
Słabicki said: “Proteins are the basic material our cells are made of; they ensure the proper development of cells. The problem starts when a protein breaks and initiates uncontrolled cell division, which can lead to the development of cancer.”
Zuzanna Kozicka added: “An important group of proteins are enzymes that control all chemical changes in the cell. The most important region of an enzyme is the active site responsible for the action of a given protein. Usually, when developing a potential drug, we look for a chemical compound that will perfectly match the shape of the active protein site we want to inhibit - just like a key matches s door lock. Such a key can stop the action of an unregulated, pathogenic protein.”
The problem, however, is that not all proteins have such active sites. Kozicka said: “When we want to neutralise a malfunctioning protein, we often simply don't have a chance of finding the right key, and many of these proteins play an important role in cancer development. That is why we need innovative approaches that will enable us to develop drugs that work in a new way.”
One of the ideas is called targeted protein degradation. It involves outsmarting the cell and forcing it to destroy the pathogenic protein by using the 'molecular machinery' responsible for getting rid of unnecessary or abnormal proteins (ubiquitin - proteasome system). For a protein to be destroyed, it must first be labelled with a special small molecule protein, ubiquitin, by an enzyme of said 'molecular machinery' called ubiquitin ligase. Targeted degradation of an undesirable protein can thus be achieved using a 'molecular glue' that combines the given protein with the ligase.
Słabicki, Kozicka and Georg Petzold are the core of an international team of scientists led by Professor Benjamin Ebert of the Dana-Farber Cancer Institute and Dr. Nicolas Thomä of the Friedrich Miescher Institute for Biomedical Research. Researchers searched for compounds that would cause protein degradation. They published the results of their search in the prestigious journal Nature, https://rdcu.be/b4BiT.
Using huge databases, the researchers found a CR8 compound acting as a molecular glue that forces ubiquitin labelling, and thus the destruction of the protein responsible for developing the disease. Researchers looked at how the amount of all proteins in the cell changes after the administration of CR8 into the cell. It turned out that the amount of all proteins remained unchanged, except for one: cyclin K. Further experiments confirmed that this protein, which probably contributes to the development of tumours, landed in the 'molecular rubbish bin', by gluing it with one of the ubiquitin ligases forced by CR8.
Kozicka said that in this situation, instead of an inhibitor, or a key inserted into the active site, we have a degrader: a molecule that makes it possible to precisely mark an undesirable, malfunctioning protein and cause its degradation in the cell. This opens up the possibility of deactivating such malfunctioning proteins that we have not been able to neutralize so far, because, for example, they do not have active sites or stable spatial structures.
The researchers have also solved the crystal structure that allows scientists to visualize proteins glued by CR8. Kozicka said: “Deciphering how these individual parts of the puzzle connect with each other allows us to better understand the functioning of molecular glues. It also allows us to slowly start thinking about 'rational designing' of this type of drugs, figuring out what a chemical compound should look like that would combine two selected protein - but this is still a very difficult task.”
Słabicki added that until now only two groups of compounds called molecular glues were known, and many pharmaceutical companies invest in this type of research, because thanks to such compounds it will be possible to treat diseases for which effective and safe medicines have not been available so far.
“We have shown that there are more such compounds, that we can and should look for them,” he said.
PAP - Science in Poland, Ewelina Krajczyńska
ekr/ agt/ kap/
tr. RL