Transposons, known as jumping genes, can 'cut themselves out' from the DNA, and then paste into its another, often precisely selected place. Scientists view them as the future of genome editing. Polish researchers have recently completed a difficult task: they described the key protein related to the action of the transposon when this is 'getting ready to jump'.
“Our work adds another brick to understanding how the protein can cut out large DNA fragments and paste them in another place,” says Professor Marcin Nowotny from the International Institute of Molecular and Cell Biology in Warsaw (IIMCB). His team - together with Joseph Peters from Cornell University (USA) - published the results of their research in the prestigious journal Molecular Cell.
Transposons are parasites in the genome. Some of them not only move, but also use the cellular machinery of organisms to multiply - explains Professor Marcin Nowotny. “Up to 50 percent of the human genome and 70 percent of the genome of some plants may come from transposons,” he says (although in most cases, these transposons are not active). Some viruses, including HIV, also come from transposons. There are also transposons, thanks to which bacteria become resistant to antibiotics. This gives a little insight into how common the 'jumping genes' are in nature.
Professor Marcin Nowotny points out that although the transposons are commonly called jumping genes, it would be more accurate to call them jumping DNA (because a transposon is not a gene, but it may contain genes).
Transposons have achieved an extraordinary evolutionary success and are very diverse. Some work by cutting themselves out from the DNA and pasting themselves in a precisely selected place of the genome. This operation resembles a text editor cut and paste command (CTRL+X and CTRL+V). If scientists want to edit the genome without any unforeseen errors - as precisely, as we want to edit texts - they should learn and use similar tricks of nature itself.
That is why they dream of developing an artificial transposon that would paste selected genes into the DNA. You can imagine that such a tool will someday save the life and health of people with genetic diseases whose DNA encodes a defective type of protein. Such people could have artificial transposons supplied to the body, which would paste themselves into the host's DNA in a safe place. And then the body itself could produce a medicine - correct protein necessary for health.
As for the DNA editing possibilities, the main tool is currently CRISPR-Cas9 (2020 Nobel Prize) called genetic scissors. This mechanism (discovered during research on bacteria) makes it possible to find a precisely selected place in the DNA and cut it. In place of the cut, genes artificially supplied to the cell can be pasted. For now, however, there is no guarantee that this will always happen, and that mutations will not form at the cut site. The CRISPR-Cas9 procedure requires improvement. We need to find a way to more precisely paste the selected gene into the cut site and reconnect the DNA.
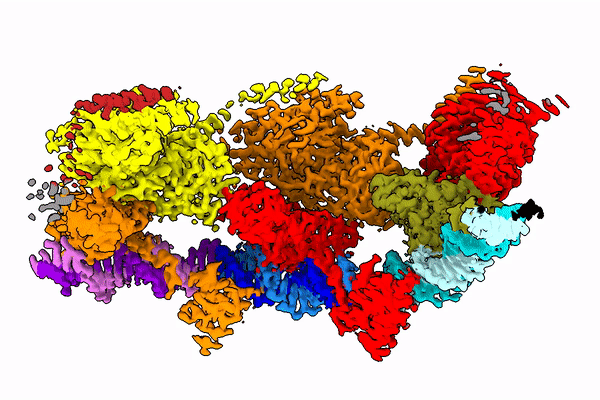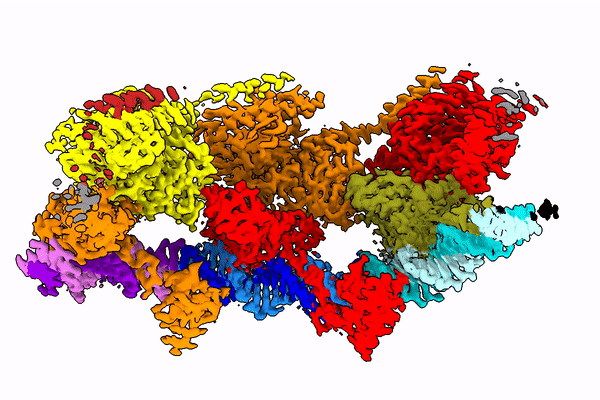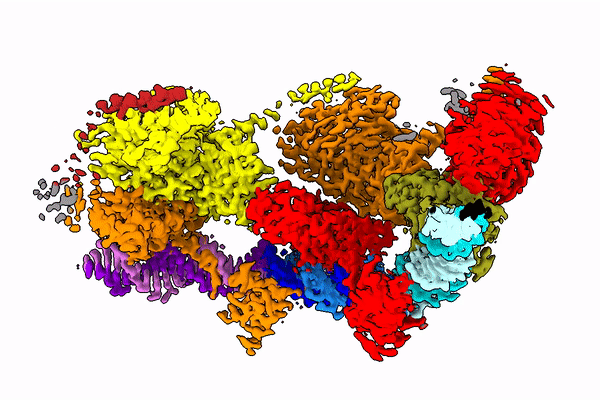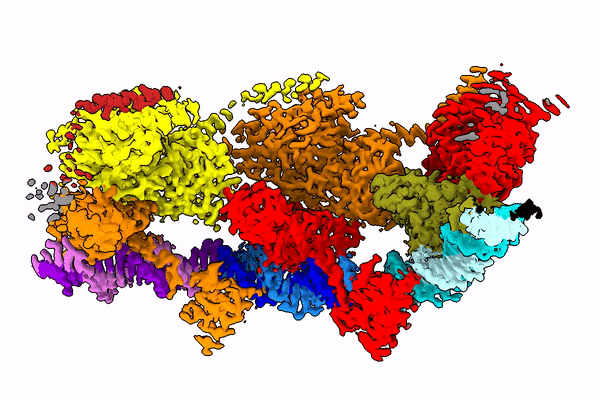
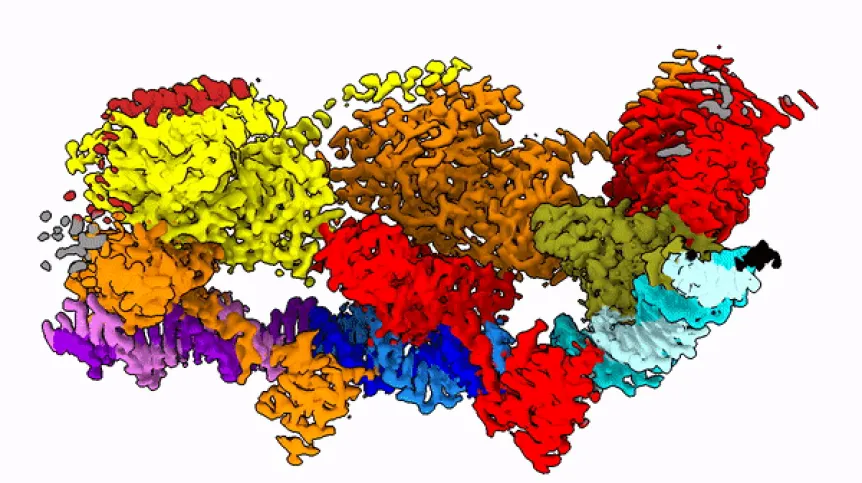
The Holy Grail in this area seems to be the Tn7 transposon obtained from bacteria. Its use seems possible, because this well-known CRISPR-Cas9 machinery is naturally coded in some of its variants it. The transposon uses it to choose the genome place where it is inserted.
One of the genes of the Tn7 transposon codes an enzyme - a transposase whose task is to cut out several thousand nucleotides (letters of the genetic code) of the transposon from one place and paste it into another.
To understand how the the enzyme performs is possible, scientists want to learn the structure of the transposase, and therefore its shape. Because its shape determines how it works in the cell. The study of such proteins is a very difficult task, because this enzyme consists of tens of thousands of atoms. In addition, it is very busy: its shape changes depending on the phase of the operation it performs.
Nowotny's team used cryoelectron microscopy (2017 Nobel Prize), which has been operated by the Krakow SOLARIS Centre for several years. This technology allows researchers to observe shadows of individual molecules frozen in motion. And based on these shadows, they can reproduce the extremely complicated 3D structures of the studied molecules.
This molecule, despite the fact that it consists of tens of thousands of atoms, is still tiny. If you change the proportions of various objects and enlarge this molecule to the size of the apple, a similarly enlarged human would have a height of 20,000 km and be able of wrapping around half of the Earth.
Professor Marcin Nowotny compares the studied protein to a string with beads. “These beads dance in space and arrange themselves quite differently depending on the phase of the enzyme jump from one place to another,” he says.
In the publication in Molecular Cell, the team shows how the protein looks at the moment when it finds its place in the DNA (after identifying the end of the DNA fragment it is supposed to cut).
Scientists from around the world are racing to show the protein model in other stages of its operation. For example, a team from Cornell University recently presented the shape of the same protein while pasting a fragment into another place.
Researchers managed to 'photograph' extremely interesting particles in a few important moments of their work. A lot of effort is still needed to learn the shape of this protein in other phases of this operation. Without this knowledge, it will be difficult to understand how this molecule works, and how to use the mechanism associated with it in practice.
PAP - Science in Poland, Ludwika Tomala
lt/ zan/ kap/
tr. RL













